DEM を扱う Shiny プログラム
動作画面例
単純表示
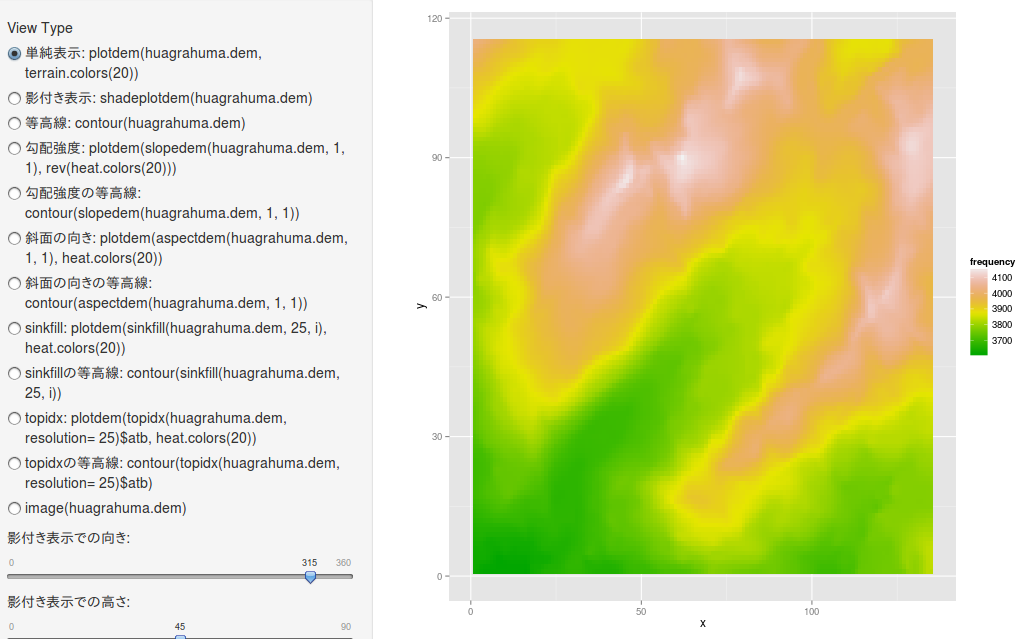
影付き表示
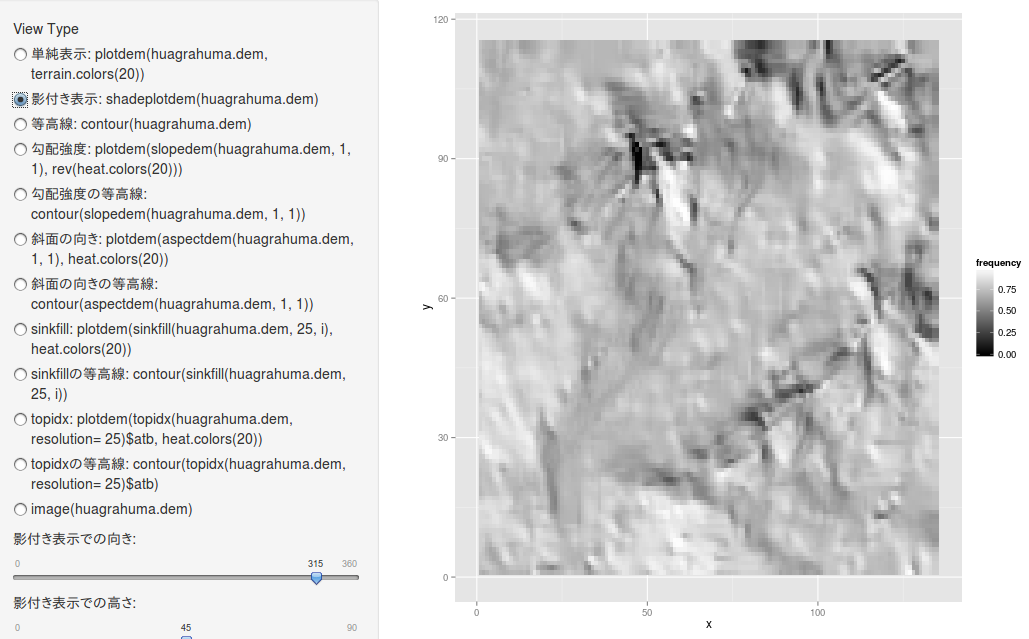
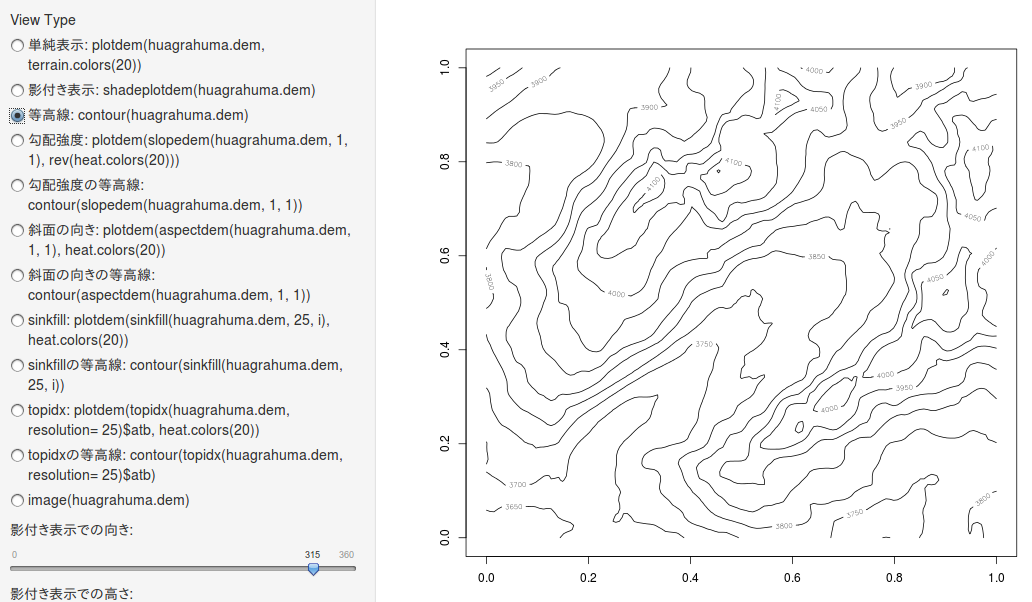
等高線
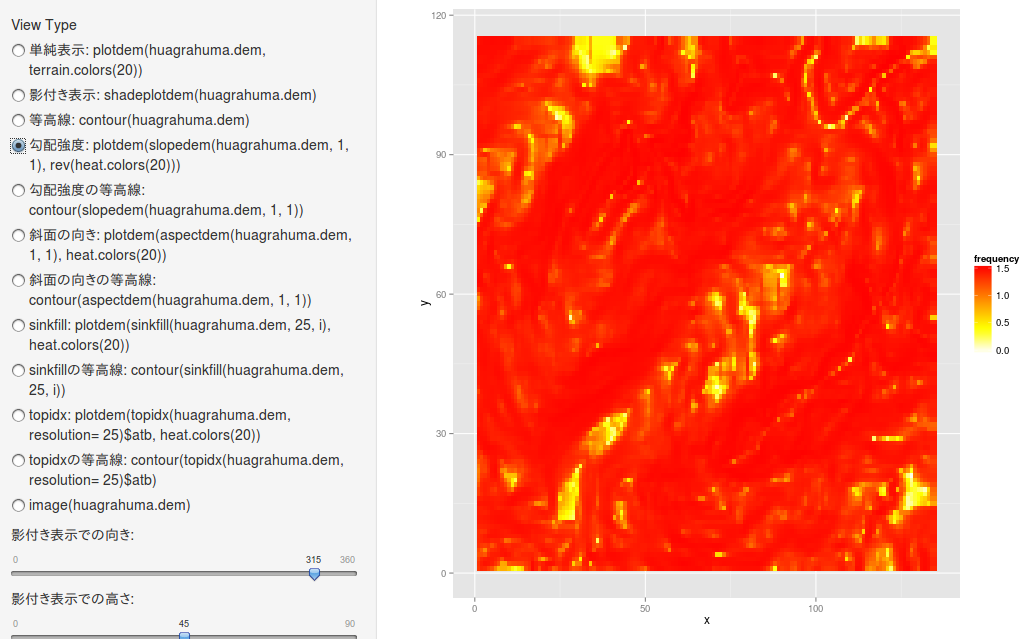
勾配強度
勾配強度の等高線
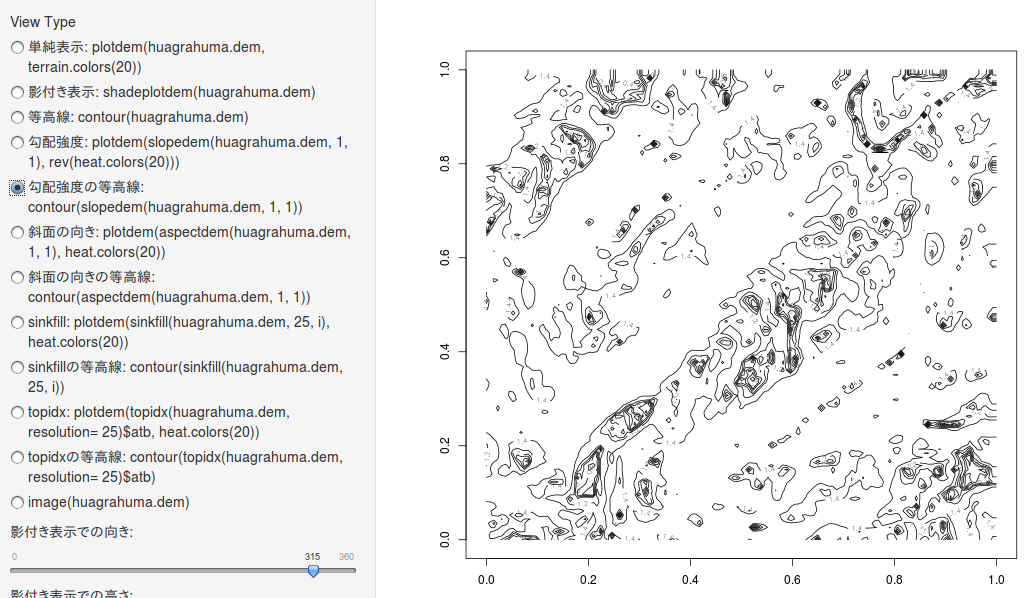
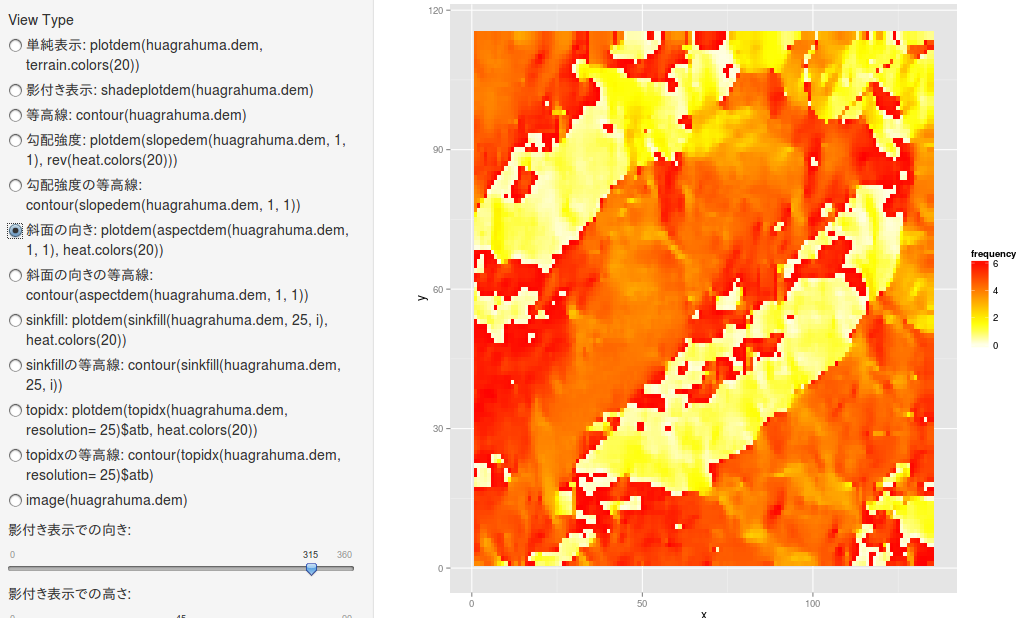
斜面の向き
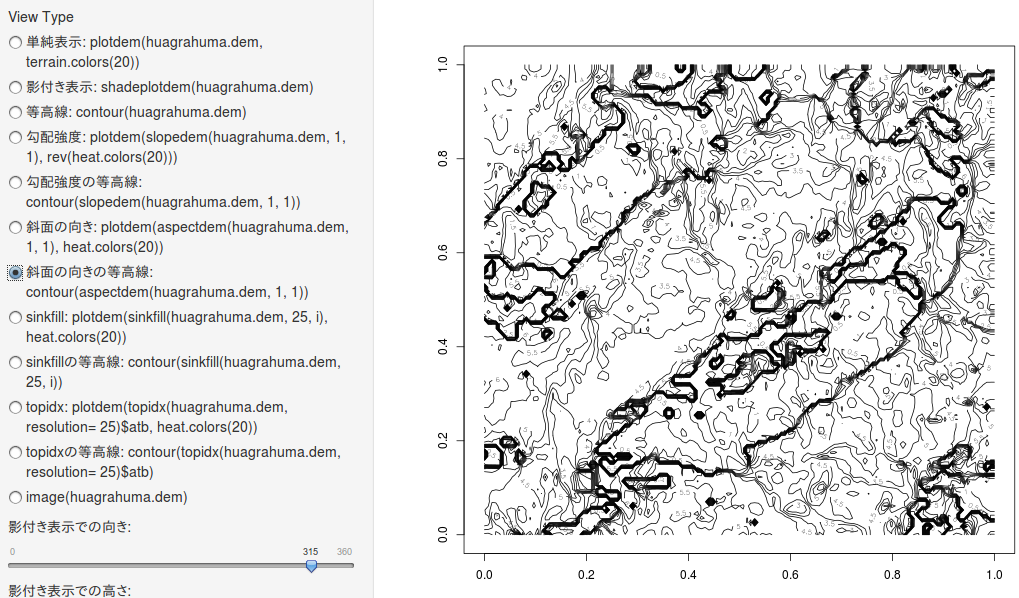
斜面の向きの等高線
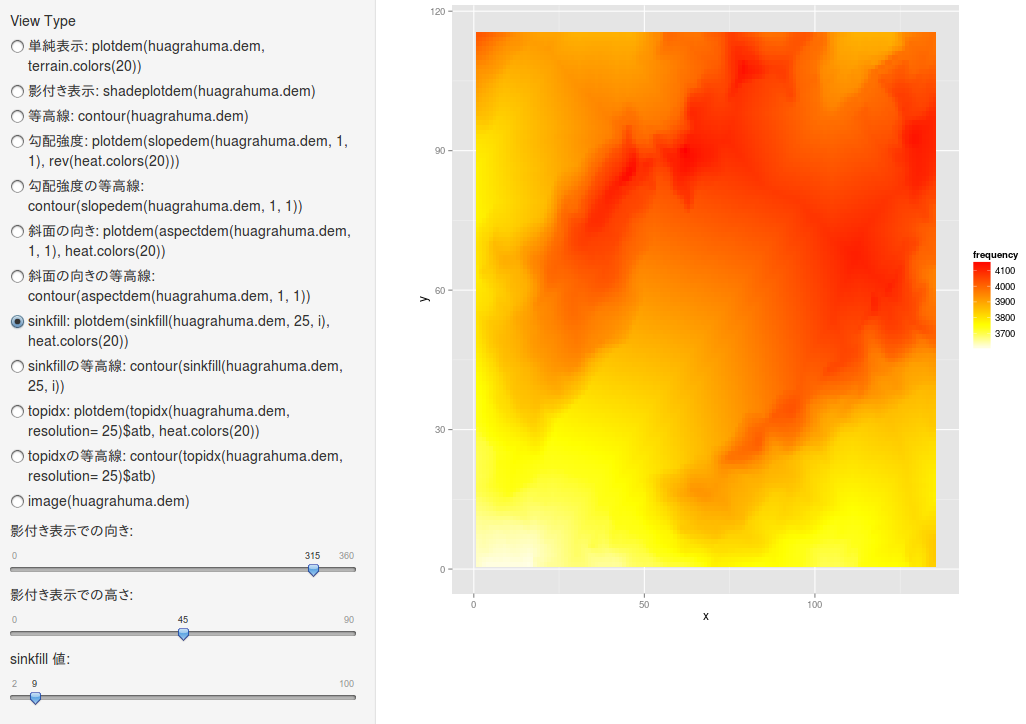
sinkfill
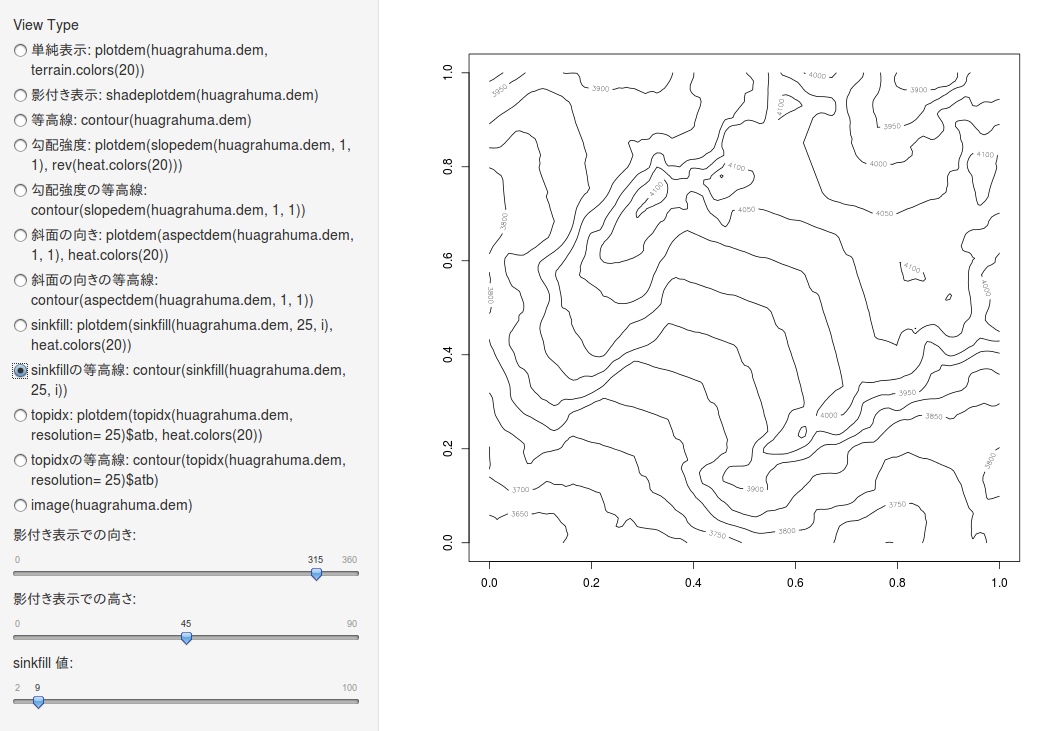
sinkfill の等高線
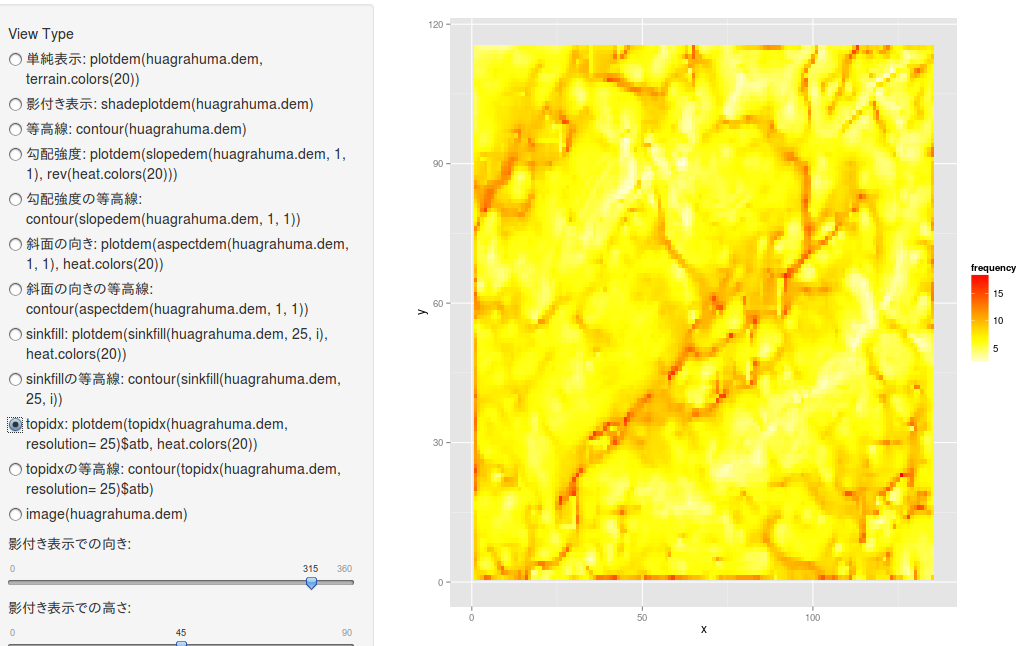
topidx
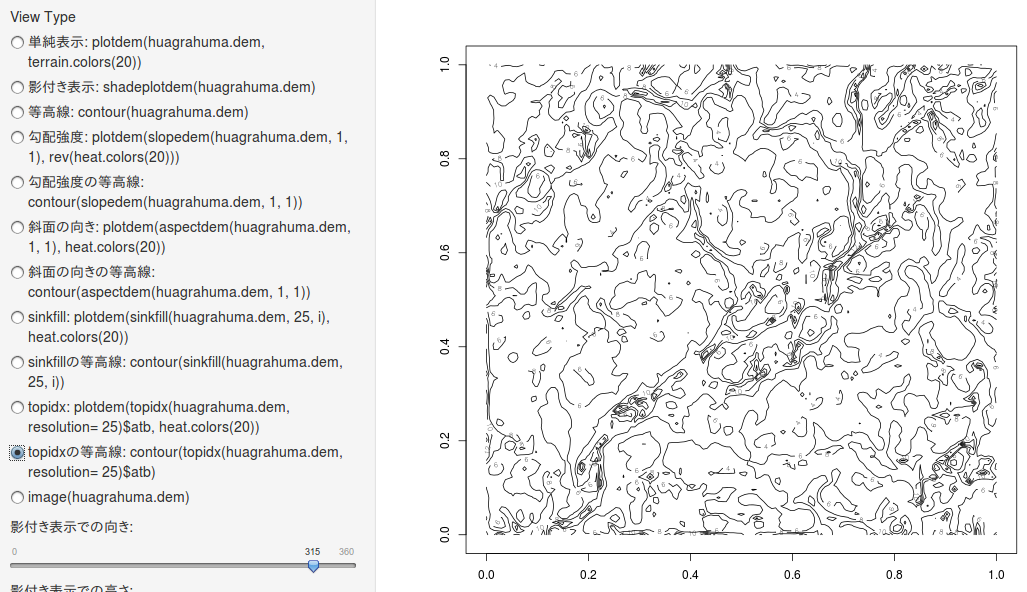
topidxの等高線
ソースコード
ui.r
require(shiny)
# Define UI for application that plots random distributions
shinyUI(pageWithSidebar(
# Application title
headerPanel("Hello!"),
# Sidebar with a slider input for number of observations
sidebarPanel(
radioButtons('vt', 'View Type',
c('単純表示: plotdem(huagrahuma.dem, terrain.colors(20))'='plot',
'影付き表示: shadeplotdem(huagrahuma.dem)'='shadeplot',
'等高線: contour(huagrahuma.dem)'='contour',
'勾配強度: plotdem(slopedem(huagrahuma.dem, 1, 1), rev(heat.colors(20)))'='slopedem',
'勾配強度の等高線: contour(slopedem(huagrahuma.dem, 1, 1))'='slopedem_contour',
'斜面の向き: plotdem(aspectdem(huagrahuma.dem, 1, 1), heat.colors(20)) '='aspectdem',
'斜面の向きの等高線: contour(aspectdem(huagrahuma.dem, 1, 1)) '='aspectdem_contour',
'sinkfill: plotdem(sinkfill(huagrahuma.dem, 25, i), heat.colors(20))'='sinkfill',
'sinkfillの等高線: contour(sinkfill(huagrahuma.dem, 25, i))'='sinkfill_contour',
'topidx: plotdem(topidx(huagrahuma.dem, resolution= 25)$atb, heat.colors(20)) '='topidx',
'topidxの等高線: contour(topidx(huagrahuma.dem, resolution= 25)$atb) '='topidx_contour',
'image(huagrahuma.dem)'='image'),
'単純表示: plotdem(huagrahuma.dem, terrain.colors(20))'),
sliderInput("dir",
"影付き表示での向き:",
min = 0,
max = 360,
value = 315),
sliderInput("a",
"影付き表示での高さ:",
min = 0,
max = 90,
value = 45),
sliderInput("i",
"sinkfill 値:",
min = 2,
max = 100,
value = 5)
),
# Show a plot of the generated distribution
mainPanel(
plotOutput("distPlot", width="100%", height="100%")
)
))
server.r
require(shiny)
require(ggplot2)
require(topmodel)
require(insol)
require(data.table)
data(huagrahuma)
data(huagrahuma.dem)
plotdem <- function(DEM, c) {
x <- rep(1:nrow(DEM), ncol(DEM)) # 1 2 3 1 2 3 1 2 3 1 2 3
y <- rep(1:ncol(DEM), each=nrow(DEM)) # 1 1 1 1 2 2 2 2 3 3 3 3
T <- data.table(x=x, y=y, val=as.numeric(DEM))
p <- ggplot(T, aes(x=x, y=y, fill=val)) + geom_tile() + scale_fill_gradientn("frequency", colours = c)
return(p)
}
slopedem <- function(DEM, dl_long, dl_lat) {
return( slope( cgrad(DEM, dl_long, dl_lat) ) )
}
aspectdem <- function(DEM, dl_long, dl_lat) {
return( aspect( cgrad(DEM, dl_long, dl_lat) ) )
}
shadeplotdem <- function(DEM, dir, a) {
cellsize=30
sv=normalvector(a, dir)
grd=cgrad(DEM, cellsize)
hsh=grd[,,1]*sv[1]+grd[,,2]*sv[2]+grd[,,3]*sv[3]
## remove negative incidence angles (self shading)
hsh=(hsh+abs(hsh))/2
sh=doshade(DEM, sv, cellsize)
p <- plotdem(hsh*sh, grey(1:100/100))
return(p)
}
shinyServer(function(input, output) {
output$distPlot <- renderPlot({
print(input$vt)
if (input$vt == 'plot') {
p <- plotdem(huagrahuma.dem, terrain.colors(20))
print(p)
}
else if (input$vt == 'shadeplot') {
p <- shadeplotdem(huagrahuma.dem, input$dir, input$a)
print(p)
}
else if (input$vt == 'contour') {
contour(huagrahuma.dem)
}
else if (input$vt == 'slopedem') {
p <- plotdem(slopedem(huagrahuma.dem, 1, 1), rev(heat.colors(20)))
print(p)
}
else if (input$vt == 'slopedem_contour') {
contour(slopedem(huagrahuma.dem, 1, 1))
}
else if (input$vt == 'aspectdem') {
p <- plotdem(aspectdem(huagrahuma.dem, 1, 1), rev(heat.colors(20)))
print(p)
}
else if (input$vt == 'aspectdem_contour') {
contour(aspectdem(huagrahuma.dem, 1, 1))
}
else if (input$vt == 'sinkfill') {
p <- plotdem(sinkfill(huagrahuma.dem, 25, input$i), rev(heat.colors(20)))
print(p)
}
else if (input$vt == 'sinkfill_contour') {
contour(sinkfill(huagrahuma.dem, 25, input$i))
}
else if (input$vt == 'topidx') {
p <- plotdem( topidx(huagrahuma.dem, resolution= 25)$atb, rev(heat.colors(20)))
print(p)
}
else if (input$vt == 'topidx_contour') {
contour( topidx(huagrahuma.dem, resolution= 25)$atb )
}
else if (input$vt == 'image') {
image(huagrahuma.dem)
}
# dist <- rnorm(n = input$n, mean = input$mean, sd = input$sd)
# hist(dist)
}, height=640, width=640)
})
run.r
require(shiny)
runApp("/var/tmp")
![[kaneko lab.]](https://www.kkaneko.jp/info/logo_png.png)